Story
MolHIT: Advancing Molecular-Graph Generation with Hierarchical Discrete Diffusion Models
Key takeaway
Researchers developed a new AI model that can generate molecular structures more efficiently for drug discovery and materials science. This advance could accelerate the pace of innovation in these critical fields.
Quick Explainer
MolHIT introduces a novel hierarchical diffusion framework for generating molecular graphs. It models molecular structure in a coarse-to-fine manner, first establishing high-level chemical identities before refining into specific atom types. This captures meaningful dependencies between atoms during the noising and denoising process. Additionally, MolHIT's decoupled atom encoding explicitly represents key properties like formal charge and aromaticity, enabling reliable generation of complex molecular motifs. The combination of the hierarchical diffusion process and the decoupled encoding allows MolHIT to effectively navigate the valid chemical space while maintaining superior structural exploration capabilities compared to previous graph-based generation approaches.
Deep Dive
Technical Deep Dive: MolHIT for Molecular-Graph Generation
Overview
MolHIT is a novel molecular graph generation framework that advances the state-of-the-art in AI-driven molecular design. It is based on a Hierarchical Discrete Diffusion Model (HDDM), which improves upon previous graph diffusion approaches by capturing meaningful chemical dependencies during the noising and denoising process. Additionally, MolHIT introduces Decoupled Atom Encoding (DAE) to better represent key molecular properties like formal charge and aromaticity.
Problem & Context
Molecular generation with AI has significant potential to accelerate materials design and drug discovery. However, generating valid and novel molecular structures is challenging due to the vast combinatorial search space. While sequence-based models can achieve high validity, they struggle to explore the chemical space beyond common patterns in the training data. Graph-based diffusion models offer improved structural exploration, but often produce chemically invalid samples.
Methodology
The key innovations in MolHIT are:
- Hierarchical Discrete Diffusion Model (HDDM): HDDM extends the discrete diffusion framework by introducing additional mid-level states that capture meaningful chemical relationships between atom types. This coarse-to-fine approach allows the model to first establish high-level chemical identities before refining into specific atom types.
- Decoupled Atom Encoding (DAE): DAE explicitly encodes atom properties like formal charge and aromaticity as primary node features. This resolves the one-to-many mapping problem in previous coarse-grained encodings, enabling reliable generation of complex molecular motifs.
- Project-and-Noise (PN) Sampling: To encourage structural diversity, MolHIT uses a custom PN sampling scheme that projects the model's denoising predictions onto the clean manifold before re-noising to the previous timestep.
Data & Experimental Setup
MolHIT is evaluated on two large-scale molecular benchmarks:
- MOSES: A dataset of 1.9M drug-like molecules, used to assess unconditional generation performance.
- GuacaMol: A more chemically diverse dataset containing compounds with formal charges, used to test the robustness of MolHIT's atom encoding.
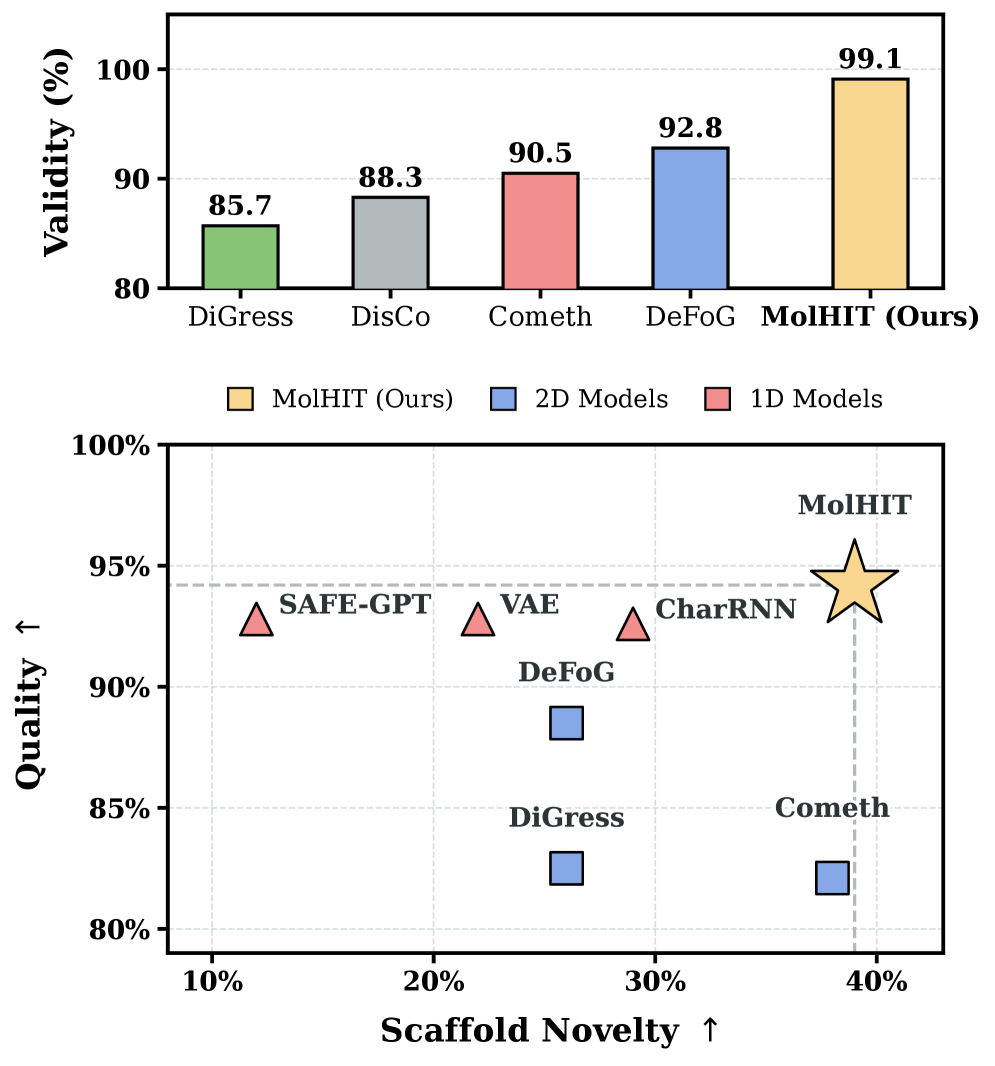
MolHIT is compared against strong 1D sequence-based baselines (e.g., VAE, Char-RNN, SAFE-GPT) as well as previous state-of-the-art graph diffusion models (e.g., DiGress, DisCo, Cometh, DeFoG).
Results
MolHIT significantly outperforms all baselines across the MOSES and GuacaMol benchmarks:
- On MOSES, MolHIT achieves a new state-of-the-art in Quality (94.2%) and Scaffold Novelty (0.39), while maintaining near-perfect Validity (99.1%).
- On the more chemically diverse GuacaMol dataset, MolHIT demonstrates robust performance, achieving the highest scores in Validity, Uniqueness, and Novel Unique.
- In conditional generation tasks, MolHIT shows strong performance in multi-property guided generation and scaffold extension, outperforming previous graph diffusion approaches.
Interpretation
The results demonstrate that MolHIT's hierarchical diffusion process and decoupled atom encoding enable it to effectively navigate the valid drug-like manifold while maintaining a superior capacity for structural exploration. The model's ability to reliably generate complex motifs and handle diverse chemical spaces suggests it is a significant advancement towards more realistic and generalizable molecular generation.
Limitations & Uncertainties
While MolHIT achieves state-of-the-art performance, the authors note that there is still room for improvement through model size scaling and further training on the GuacaMol dataset. Additionally, the authors did not explore the application of MolHIT's HDDM framework to other domains like language modeling or image generation.
What Comes Next
Promising future directions include:
- Applying the HDDM framework to language models and image generation tasks.
- Incorporating more advanced molecular representations, such as those capturing functional groups or 3D structure.
- Exploring the use of MolHIT for protein design and other molecular engineering applications beyond small-molecule generation.
- Investigating the integration of MolHIT with other reinforcement learning or optimization techniques for targeted molecular design.