Story
Guide-Guard: Off-Target Predicting in CRISPR Applications
Key takeaway
A new AI-powered tool can help predict and avoid unintended genetic changes when using CRISPR gene editing, potentially making this powerful biotech tool safer and more precise for medical and agricultural applications.
Quick Explainer
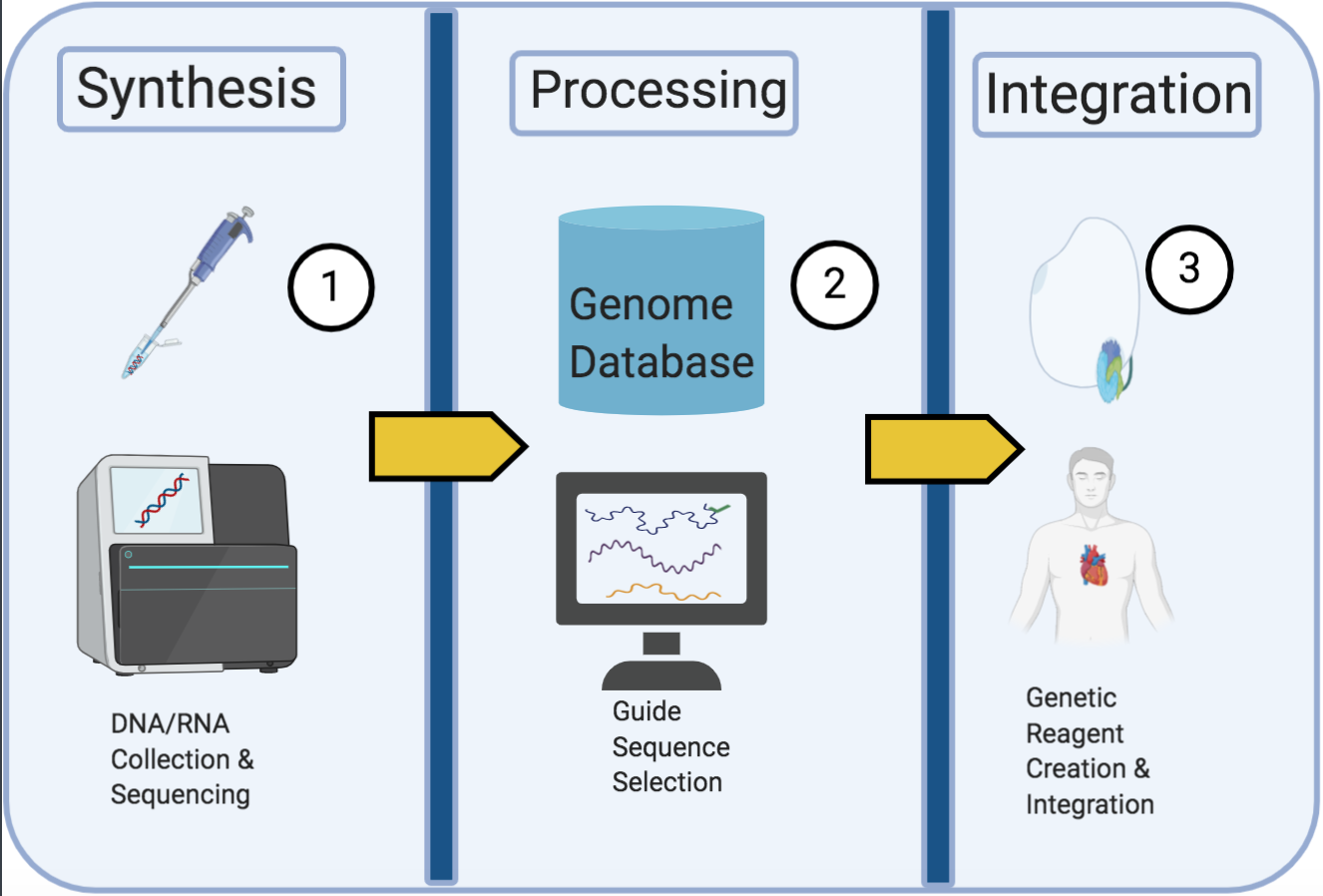
Guide-Guard is a machine learning-based tool that aims to predict the safety and off-target behavior of guide RNA sequences used in CRISPR gene editing. It analyzes how the location and type of mismatches in the guide RNA impact the binding potential of the Cas13 enzyme. By incorporating this domain knowledge, Guide-Guard's convolutional neural network can efficiently classify guide RNA sequences as safe or unsafe for use, helping to mitigate potential safety issues or misuse of CRISPR technology.
Deep Dive
Technical Deep Dive: Guide-Guard: Off-Target Predicting in CRISPR Applications
Overview
The paper presents a machine learning-based solution called "Guide-Guard" to predict the safety and off-target behavior of guide RNA (gRNA) sequences used in CRISPR gene editing. The key contributions are:
- Analysis of how mismatch location and nucleotide type impact CRISPR Cas13 binding potential.
- Development of the Guide-Guard convolutional neural network model to classify gRNA sequences as safe or unsafe.
- Validation of Guide-Guard's 84% accuracy on a real-world CRISPR Cas13 dataset.
- Discussion of cyberbiosecurity risks and Guide-Guard's potential to mitigate them by vetting gRNA before use.
Methodology
- The researchers used a dataset from prior work that included intentionally induced mismatches in gRNA sequences to study off-target effects.
- They observed that mismatch location, especially at the 5th and 18th nucleotides, and nucleotide type (U vs G/C) had significant impact on Cas13 binding potential.
- They incorporated this domain knowledge into the design of their convolutional neural network model, Guide-Guard, which takes as input the gRNA sequence and the reverse-complement of the target sequence.
- Guide-Guard classifies gRNA sequences into 8 equally-distributed classes based on predicted off-target binding, with the top class considered safe for use.
Results
- Guide-Guard achieved 84% overall accuracy in classifying gRNA sequences as safe or unsafe.
- It had higher accuracy (85.51%) on perfect match gRNA compared to mismatched gRNA (77.50%).
- The model's ROC curve had an area under the curve of 0.839, indicating strong predictive performance.
- Guide-Guard is computationally efficient, taking only 0.00055 seconds on average to process a single input on a 2011 Macbook Pro.
Interpretation
- The mismatch analysis provides insight into the structural factors governing Cas13 binding, which the researchers leveraged to improve Guide-Guard's performance.
- Guide-Guard's high accuracy and efficiency make it a promising tool to vet gRNA sequences before use, preventing potential safety issues or misuse.
- Integrating Guide-Guard into gene editing workflows, gene banks, or synthesis pipelines could improve the overall cyberbiosecurity of CRISPR applications.
Limitations & Uncertainties
- The dataset was limited to 3 target genes, so the generalizability to a wider range of genes is unclear.
- The paper does not explore whether Guide-Guard's performance holds for other CRISPR systems like Cas9 beyond Cas13.
- The study does not address the potential for adversarial attacks on the model itself, which could undermine its security guarantees.
What Comes Next
- Expanding the training dataset to cover a broader range of genes and CRISPR systems.
- Investigating techniques to make Guide-Guard more robust to potential adversarial attacks.
- Integrating Guide-Guard into real-world gene editing pipelines and measuring its impact on safety and productivity.
- Exploring extensions of the approach to other stages of the gene editing workflow beyond gRNA selection.
Sources: