Story
Enhanced Diffusion Sampling: Efficient Rare Event Sampling and Free Energy Calculation with Diffusion Models
Key takeaway
Researchers developed a new algorithm that can efficiently simulate rare molecular events, like chemical reactions or protein folding. This could lead to faster and more accurate computer models of complex biological processes.
Quick Explainer
The core idea of Enhanced Diffusion Sampling is to integrate classical enhanced sampling techniques, such as umbrella sampling and metadynamics, into diffusion-based equilibrium samplers. This enables efficient computation of free energies and rare-state observables for complex molecular processes. The approach involves a steering procedure that generates biased ensembles from a pretrained diffusion model, followed by exact reweighting to recover unbiased expectation values. This integration of classical and diffusion-based methods addresses the slow mixing problem of molecular dynamics simulations and the rare state problem, which limits the ability of diffusion models to estimate observables dominated by low-probability regions.
Deep Dive
Technical Deep Dive: Enhanced Diffusion Sampling for Rare Event Estimation
Overview
This paper introduces an enhanced sampling framework that integrates classical bias-and-reweighting methods into diffusion-based equilibrium samplers. This enables efficient computation of free energies and rare-state observables for complex biomolecular processes, closing a key gap left by recent advances in generative diffusion models.
Problem & Context
Molecular dynamics (MD) simulation faces two key bottlenecks:
- Slow mixing problem: MD produces time-correlated trajectories, leading to slow exploration and slow convergence of expectation values.
- Rare state problem: Even with independent samples, it is prohibitive to sample states with small equilibrium probabilities, such as unfolded protein conformations.
Traditional enhanced sampling methods can address the rare state problem, but are limited by the slow mixing of MD. Recently, diffusion-based equilibrium samplers have emerged that generate independent samples, tackling the slow mixing issue. However, these models still struggle to estimate observables dominated by low-probability regions.
Methodology
The authors' key insight is to integrate classical enhanced sampling techniques (umbrella sampling, metadynamics, free energy differences) into the diffusion model framework. This is achieved through a steering procedure that generates biased ensembles from a pretrained diffusion model, followed by exact reweighting to recover unbiased expectation values.
The three main methods introduced are:
- UmbrellaDiff: Adapts umbrella sampling to diffusion models, enabling efficient exploration of free energy landscapes without the need for kinetically connected windows.
- MetaDiff: Reformulates metadynamics for diffusion models, allowing online updates of the bias potential and unbiased free energy estimation at any point.
- ΔG-Diff: Computes free energy differences between two states (e.g. folded and unfolded protein) by steering the diffusion model with a biasing potential.
Data & Experimental Setup
The authors test their methods on a range of biomolecular systems, primarily using the pretrained BioEmu diffusion model:
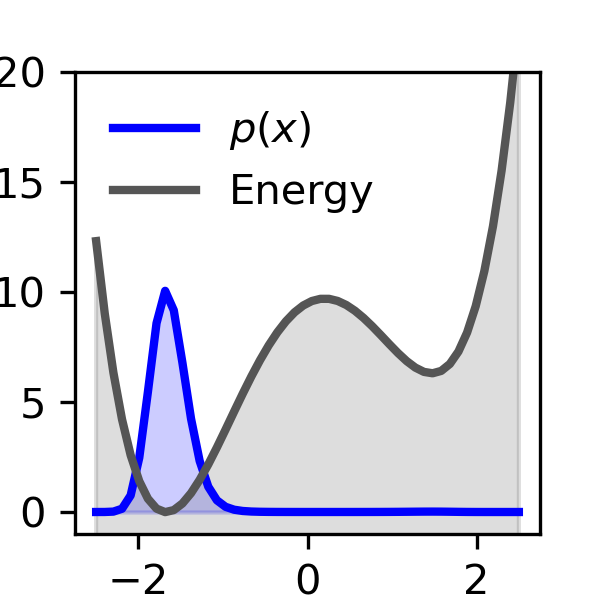
- Toy double-well potentials to illustrate the core concepts
- 18 proteins from the ProThermDB database, ranging from 76 to 372 residues, with predicted folding free energies from 1.1 to 5.7 kcal/mol
The key evaluation metric is the minimum number of samples required to achieve convergence of the free energy estimate within 1 kcal/mol.
Results
- Toy double-well potentials: Enhanced diffusion sampling achieves convergence with 10-100 samples, compared to an exponential increase in samples required for unbiased sampling as the free energy difference grows.
- Protein folding free energies:
- Steered sampling eliminates "catastrophic failures" (no unfolded samples) at small sample sizes, whereas unbiased sampling requires orders of magnitude more particles for very stable proteins.
- Steered sampling achieves sub-kcal/mol accuracy at 100-1,000 samples, while unbiased sampling requires exponentially more samples as the free energy difference increases.
- The advantage of steered sampling becomes more pronounced for proteins with higher folding free energies (> 3.2 kcal/mol).
Interpretation
The authors show that integrating classical enhanced sampling techniques into diffusion-based equilibrium samplers can dramatically improve the efficiency of estimating rare-event observables, like protein folding free energies. This closes a key gap left by recent progress in generative diffusion models, which excel at generating independent samples but struggle to estimate observables dominated by low-probability regions.
Limitations & Uncertainties
- The approach relies on having an accurate pretrained diffusion model for the molecular system of interest. Any model mismatch will propagate into the reweighted estimates.
- Weight degeneracy can arise if the biased ensembles have insufficient overlap, requiring careful bias design and diagnostics.
- The focus is on equilibrium properties; extending these ideas to dynamical observables will require additional methodological developments.
What Comes Next
The authors suggest several promising future directions:
- Combining the steering framework with adaptive schemes for learning optimal reaction coordinates or bias potentials.
- Leveraging energy-based diffusion models that allow direct evaluation of the energy function, enabling tighter estimator control and new forms of biasing.
- Unifying learned transport models (e.g. MDGen, Timewarp) with thermodynamically consistent sampling via Metropolis-Hastings or path reweighting.
- Applying the enhanced diffusion sampling approach to a broader range of application areas beyond biomolecular systems, such as materials science and condensed-phase chemistry.