Story
Systematic Evaluation of Single-Cell Foundation Model Interpretability Reveals Attention Captures Co-Expression Rather Than Unique Regulatory Signal
Key takeaway
Researchers found that AI models used to analyze single-cell data may not capture regulatory signals, but instead just track gene co-expression patterns. This means these models may not provide meaningful biological insights.
Quick Explainer
The paper presents a systematic framework to evaluate the interpretability of single-cell foundation models, focusing on scGPT and Geneformer. The key insight is that while these models encode structured biological information in their attention patterns, this information does not provide incremental value for predicting the outcomes of genetic perturbations. Instead, simple gene-level features like expression and dropout rates are more predictive. The authors introduce a constructive approach called Cell-State Stratified Interpretability to enhance attention-based gene regulatory network recovery, addressing a specific scaling failure in attention mechanisms. The core contribution is a comprehensive evaluation pipeline that disentangles the mechanistic interpretability of these models from their overall predictive capacity.
Deep Dive
Technical Deep Dive: Attention Captures Co-Expression, Not Regulatory Signal
Overview
This paper presents a systematic evaluation framework to assess the mechanistic interpretability of single-cell foundation models, with a focus on scGPT and Geneformer. The key findings are:
- Attention patterns in these models encode structured biological information, with early layers capturing protein-protein interactions and later layers capturing transcriptional regulation.
- However, this structural information provides no incremental value for predicting the outcomes of genetic perturbations. Simple gene-level features like variance, mean expression, and dropout rate outperform both attention-derived and correlation-based edge scores.
- The attention-correlation relationship is context-dependent, with attention outperforming correlation in some cell types (RPE1) but not others (K562). However, the underlying pattern is consistent - gene-level features dominate perturbation prediction.
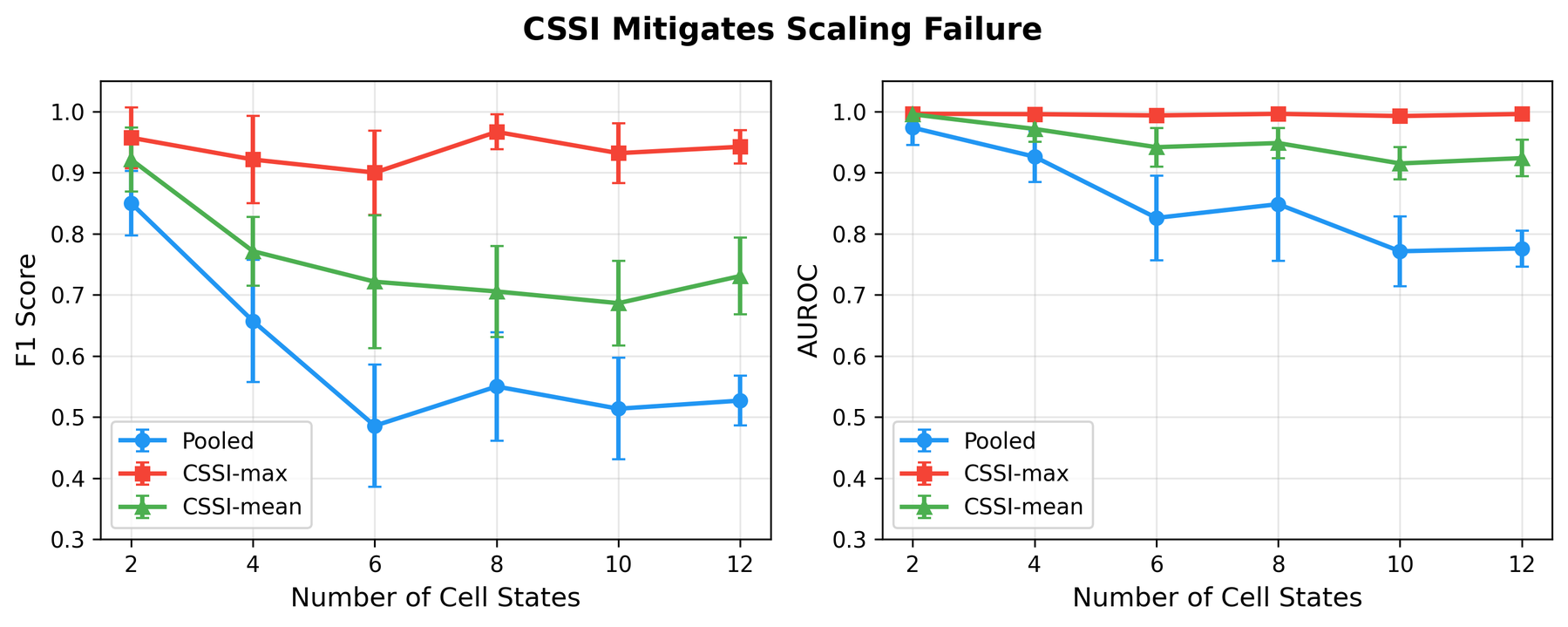
- The authors introduce Cell-State Stratified Interpretability (CSSI) as a constructive approach to improve attention-based gene regulatory network recovery, addressing an attention-specific scaling failure.
Methodology
The authors designed a multi-faceted evaluation framework with five key components:
- Trivial-baseline comparison: Evaluating whether pairwise edge scores outperform simple gene-level features.
- Conditional incremental-value testing: Assessing whether edge scores add predictive value beyond gene-level features under increasingly challenging generalization protocols.
- Expression residualization and propensity matching: Isolating edge-specific signal by removing gene-level confounds.
- Causal ablation with fidelity diagnostics: Testing whether "regulatory" attention heads causally contribute to perturbation prediction.
- Cross-context replication: Evaluating generalization across cell types and perturbation modalities.
The framework also includes a "boundary condition" tier to characterize the broader evaluation landscape, covering topics like cross-species transfer, pseudotime directionality, batch effects, and uncertainty calibration.
Results
The key findings from this extensive evaluation are:
- Attention patterns encode layer-specific biological structure, but this information provides no incremental value for perturbation prediction.
- Trivial gene-level features like variance, mean expression, and dropout rate outperform both attention-derived and correlation-based edge scores for predicting perturbation outcomes.
- Adding pairwise edge scores, whether from attention or correlation, provides zero incremental predictive value beyond gene-level features.
- Causal ablation of "regulatory" attention heads produces no degradation in perturbation prediction, while random ablation does cause significant drops.
- The attention-correlation relationship is context-dependent, with attention outperforming correlation in RPE1 cells but underperforming in K562 CRISPRa.
- However, the underlying pattern of gene-level dominance generalizes across cell types.
Limitations and Uncertainties
- Causal ablation was only tested on Geneformer, not scGPT, due to architectural differences.
- The definitive "positive control" demonstrating recovery of known causal regulatory structure is still an open challenge for the field.
- The generalization of findings is limited to the specific cell types, species, and model architectures evaluated.
- Reference database circularity affects all gene regulatory network evaluation, though the authors show their conclusions are robust to restricting to well-characterized regulatory interactions.
Implications and Next Steps
The key implication is that attention patterns in these single-cell foundation models do not directly capture causal regulatory relationships, despite being commonly interpreted as doing so. The value of these models lies in their overall predictive capacity, not in the mechanistic interpretability of their attention weights.
The authors propose three promising directions for future work:
- Intervention-aware pretraining on perturbation data to embed causal structure.
- Hybrid architectures using foundation model embeddings as inputs to dedicated GRN inference modules.
- Deploying CSSI-enhanced pipelines with conformal prediction sets for improved edge recoverability and uncertainty quantification.
The central challenge remains bridging the gap between edge recoverability against curated references and predictive validity for perturbation outcomes.