Story
Probability-Invariant Random Walk Learning on Gyral Folding-Based Cortical Similarity Networks for Alzheimer's and Lewy Body Dementia Diagnosis
Key takeaway
Researchers developed an algorithm that analyzes brain networks to help diagnose Alzheimer's and Lewy body dementia, which have similar symptoms but require different treatments.
Quick Explainer
The key idea is to model individualized brain networks derived from cortical gyral folding patterns, rather than using standard brain atlases. This approach better captures disease-specific anatomical variations. The framework, called PaIRWaL, uses probability-invariant random walks to encode the variable-sized, irregularly-structured networks, preserving their structural and anatomical properties. The encoded walk representations are then processed by a neural network to classify Alzheimer's disease and Lewy body dementia. This innovative network representation and learning strategy, combined with the use of biologically-grounded cortical landmarks, demonstrates consistent improvements over existing gyral folding network and atlas-based approaches for differential diagnosis.
Deep Dive
Technical Deep Dive: Probability-Invariant Random Walk Learning on Gyral Folding Networks for Dementia Diagnosis
Overview
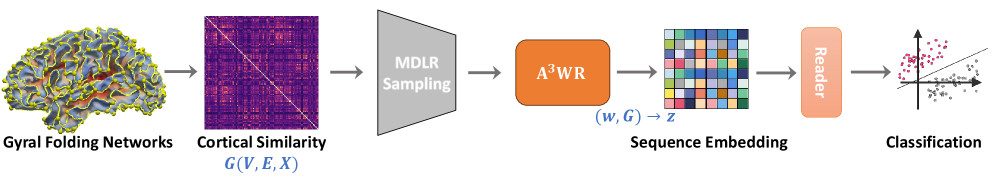
This work proposes a novel framework called PaIRWaL (Probability-Invariant Random-Walk Learning) for classifying Alzheimer's disease (AD) and Lewy body dementia (LBD) using individualized brain network models derived from cortical gyral folding patterns. Key innovations include:
- Constructing cortical similarity networks from local morphometric features of three-hinge gyri (3HGs), a biologically-grounded cortical landmark
- Modeling these variable-size, irregularly-structured networks using probability-invariant random walk sampling and anonymized walk encoding
- Demonstrating consistent improvements over existing gyral folding network and atlas-based graph learning approaches for AD/LBD diagnosis
Problem & Context
- AD and LBD are major neurodegenerative disorders with overlapping clinical symptoms, making accurate differential diagnosis crucial
- Brain network analysis using structural and functional connectivity is a promising approach, but standard atlas-based parcellations may obscure disease-sensitive individual anatomy
- Gyral folding-based networks can better capture disease-related patterns, but suffer from high inter-subject variability in node correspondence and topology
Methodology
- Cortical Similarity Network Construction:
- 3HG landmarks were automatically extracted from T1-weighted MRI scans
- Cortical morphometric features were extracted within 5-ring neighborhoods around each 3HG
- Similarity networks were built using Kullback-Leibler divergence between multivariate feature distributions
- Probability-Invariant Random Walk Learning:
- Random walks were sampled on the cortical networks using a conductance-based scheme that preserves isomorphism invariance
- Each walk was encoded into an anonymized sequence capturing structural transitions, neighborhood relations, and anatomical region attributes
- A reader neural network processed the walk encodings to output graph-level predictions
- Architecture and Training:
- Random walk length
ℓ=64,K=8walks sampled per graph for training,K=64for inference - Reader network: 2-layer MLP with 128 hidden units, trained using cross-entropy loss
- Random walk length
Data & Experimental Setup
- Cohort: 303 T1-weighted MRI scans, including 108 cognitively normal controls, 90 AD, and 105 LBD subjects
- Data preprocessing: cortical surface reconstruction, spherical mapping, 3HG extraction using automated pipeline
- Cross-validation: 10-fold splits for 4 classification tasks (AD/LBD/CN, AD vs CN, LBD vs CN, AD vs LBD)
- Evaluation metrics: accuracy, sensitivity, specificity, AUC, Matthews correlation coefficient
Results
- PaIRWaL consistently outperformed both gyral folding network baselines (GraphStat, GNN models) and atlas-based approaches (BrainNetCNN, BNT, CPSSM) across all tasks.
- On the challenging AD vs. LBD binary classification, PaIRWaL achieved substantial gains in both sensitivity and specificity compared to prior methods.
- Ablation studies confirmed the importance of probability-invariant random walks, anonymization, and anatomical attribute encoding for robust performance.
Interpretation
- The probability-invariant random walk representation effectively handles the high variability in cortical folding patterns, enabling reliable network-level disease classification.
- The anatomically-informed encoding captures meaningful structural relationships beyond just sequential transitions, providing interpretable insights into disease-relevant brain regions.
- The efficient sampling and aggregation strategy ensures computational feasibility while retaining strong predictive power.
Limitations & Uncertainties
- The study was conducted on a single clinical cohort, so generalization to other datasets remains to be evaluated.
- The specific choices of morphometric features, neighborhood size, and network sparsity thresholds were not extensively explored.
- While the work demonstrates improved AD/LBD classification, its impact on early detection and prognosis prediction is not assessed.
What Comes Next
- Validating the framework on independent cohorts and across different neuroimaging modalities (e.g., diffusion MRI, functional MRI)
- Extending the method to jointly model multimodal brain networks and investigate their complementary contributions
- Exploring ways to further enhance the interpretability of the learned representations, such as identifying specific folding patterns or network motifs associated with each disease