Story
Beyond Message Passing: A Symbolic Alternative for Expressive and Interpretable Graph Learning
Key takeaway
Researchers developed a new graph learning approach that aims to be more transparent and interpretable than existing black-box methods, which could improve trust in important applications like drug discovery.
Quick Explainer
SymGraph is a symbolic framework for graph learning that aims to overcome the limitations of standard Graph Neural Networks. It replaces continuous message passing with discrete structural hashing and topological role-based aggregation, enabling it to theoretically surpass the expressive power of existing GNNs. SymGraph constructs logical programs that fuse a node's topological signature with learned semantic states, providing higher expressiveness than standard message-passing approaches. By representing graphs as "bags of predicates" rather than opaque embeddings, SymGraph enables transparent, quantitative global reasoning through hierarchical logical rules, offering superior semantic granularity in its explanations compared to existing self-explainable GNN methods.
Deep Dive
Technical Deep Dive: Beyond Message Passing for Expressive and Interpretable Graph Learning
Overview
This paper introduces SymGraph, a symbolic framework for graph learning that aims to overcome key limitations of standard Graph Neural Networks (GNNs) in terms of expressiveness and interpretability. SymGraph replaces continuous message passing with discrete structural hashing and topological role-based aggregation, enabling it to theoretically surpass the Weisfeiler-Lehman (1-WL) expressivity barrier of standard GNNs. The key innovations are:
- Structural Hashing: SymGraph uses discrete structural hashing to capture the topological skeleton of a node's local neighborhood, avoiding the information loss inherent in continuous message passing.
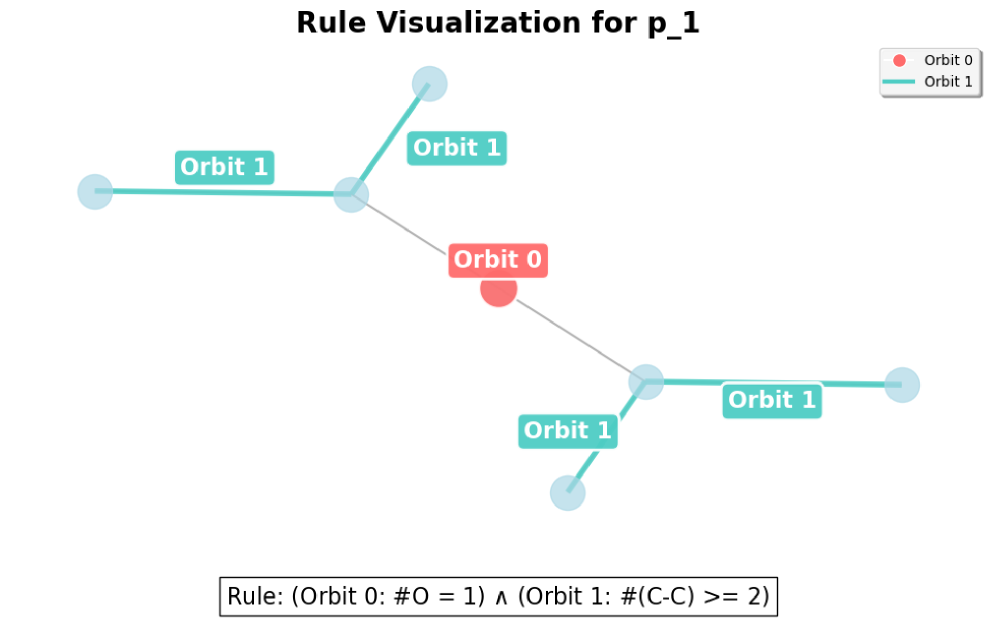
- Topological Role-based Aggregation: SymGraph partitions a node's neighborhood into orbits (equivalence classes of structurally equivalent nodes) and aggregates features accordingly, preventing "feature mixing" across distinct structural roles.
- Structure-Aware Predicates: SymGraph defines composite predicates that fuse the topological signature with learned semantic states, providing strictly higher expressiveness than standard MPNNs.
- Predicate Counting: SymGraph represents graphs as "bags of predicates" rather than opaque embeddings, enabling transparent, quantitative global reasoning through hierarchical logical rules.
Methodology
- SymGraph leverages discrete structural hashing and topological role-based aggregation to construct logical programs that theoretically surpass the 1-WL expressivity barrier.
- It employs an efficient combinatorial evolutionary search over candidate symbolic programs, overcoming the overhead of differentiable optimization in standard self-explainable GNNs.
- SymGraph's symbolic nature allows it to enforce strict structural constraints on nodes and edges, achieving finer semantic granularity in its explanations compared to existing methods.
Data & Experimental Setup
- SymGraph is evaluated on a diverse set of graph and node classification benchmarks, including both standard XAI datasets (Ba2Motifs, BAMultiShapes) and real-world molecular tasks (Mutagenicity, BBBP).
- It is benchmarked against state-of-the-art self-explainable GNNs as well as standard message-passing GNN baselines.
- Experiments are conducted on an Ubuntu machine with an Intel i7-12700K CPU and an NVIDIA GeForce RTX 5090 GPU.
Results
- SymGraph consistently outperforms state-of-the-art self-explainable GNNs in classification accuracy, achieving up to 6% improvement on molecular datasets.
- Notably, SymGraph -RF outperforms the theoretically equivalent GIN model across all benchmarks, demonstrating its ability to capture higher-order topological patterns beyond the 1-WL limit.
- In terms of efficiency, SymGraph achieves 10x to 100x speedups in training time compared to self-explainable baselines, requiring only CPU computation.
Interpretation
- SymGraph's symbolic nature allows it to generate rules that align closely with established chemical knowledge, such as SMARTS patterns used in Structure-Activity Relationship (SAR) analysis.
- Compared to existing self-explainable methods, SymGraph's explanations provide superior semantic granularity, identifying specific functional groups and structural motifs that directly explain molecular properties.
- This tight coupling between the model's logic and scientific domain knowledge offers great potential for accelerating scientific discovery and enabling truly interpretable AI systems in high-stakes domains like drug discovery.
Limitations & Uncertainties
- While SymGraph demonstrates strong performance on the evaluated benchmarks, its applicability to larger, more complex real-world graphs remains an open question that requires further investigation.
- The paper does not provide a comprehensive analysis of the computational complexity or scalability of the evolutionary search procedure, which could be a limiting factor for certain applications.
- The alignment between SymGraph's rules and SMARTS patterns is discussed conceptually, but a detailed, quantitative evaluation of this correspondence is not presented.
What Comes Next
- Future work will focus on applying SymGraph to larger, more diverse biochemistry datasets to further evaluate its potential for discovering novel Structure–Activity Relationships.
- Improvements to the evolutionary search strategy and the integration with RDKit for seamless chemical reasoning could enhance the framework's practicality and usability for domain experts.
- Exploring the generalization capabilities of SymGraph's symbolic rules, particularly in the context of transferring knowledge across related chemical tasks, is another promising direction for future research.